UDL syntax highlighting: non-delimited keywords (for nucleotide sequences)
-
Hello, @n-iels, @peterjones and All,
Edited 2019-11-21 : Sorry, @n-iels, as I’m French, I used the wrong term
ADN, instead ofDNA!Of course the Peter’s solution, using @ekopalypse Python code, is quite robust but, may be, this macro, below, could help you to study your ADN sequences ;-)). Roughly, this macro :
-
First, execute the command
Search > Unmark All > Clear All Styles -
Then, write the
ATGCupper-cased string, at beginning of the current line -
Mark, successively the
4lettersA,T,GandCwith, respectively, the styles1,2,3, and5 -
Re-select the
ATGCstring, at beginning of current line -
Delete the
ATGCstring, leaving the contents of current line untouched !
=> As a result, all the
ADNsequences of the current file should be highlighted, with the4different styles ;-))
So :
-
Start Notepad++
-
Open the
shortcuts.xmlconfiguration file, generally located in%AppData%\Roaming\Notepad++for an installed N++ version or in the same folder than the Notepad++.exe file, in case of a local installation with a zip or 7z package -
Insert the code, below, in the
<Macros>......</Macrossection ofshortcuts.xml
<Macro name="ADN Test" Ctrl="no" Alt="no" Shift="no" Key="0"> <Action type="2" message="0" wParam="43032" lParam="0" sParam="" /> <!-- DELETE ALL styles --> <Action type="0" message="2453" wParam="0" lParam="0" sParam="" /> <!-- Go to START of line --> <Action type="1" message="2170" wParam="0" lParam="0" sParam="A" /> <!-- Write the letter A --> <Action type="1" message="2170" wParam="0" lParam="0" sParam="T" /> <!-- Write the letter T --> <Action type="1" message="2170" wParam="0" lParam="0" sParam="G" /> <!-- Write the letter G --> <Action type="1" message="2170" wParam="0" lParam="0" sParam="C" /> <!-- Write the letter C --> <Action type="0" message="2453" wParam="0" lParam="0" sParam="" /> <!-- Go to START of line --> <Action type="0" message="2307" wParam="0" lParam="0" sParam="" /> <!-- Select the NEXT char --> <Action type="2" message="0" wParam="43022" lParam="0" sParam="" /> <!-- Apply the 1st STYLE --> <Action type="0" message="2306" wParam="0" lParam="0" sParam="" /> <!-- Hit the RIGHT key --> <Action type="0" message="2307" wParam="0" lParam="0" sParam="" /> <!-- Select the NEXT char --> <Action type="2" message="0" wParam="43024" lParam="0" sParam="" /> <!-- Apply the 2nd STYLE --> <Action type="0" message="2306" wParam="0" lParam="0" sParam="" /> <!-- Hit the RIGHT key --> <Action type="0" message="2307" wParam="0" lParam="0" sParam="" /> <!-- Select the NEXT char --> <Action type="2" message="0" wParam="43026" lParam="0" sParam="" /> <!-- Apply the 3rd STYLE --> <Action type="0" message="2306" wParam="0" lParam="0" sParam="" /> <!-- Hit the RIGHT key --> <Action type="0" message="2307" wParam="0" lParam="0" sParam="" /> <!-- Select the NEXT char --> <Action type="2" message="0" wParam="43030" lParam="0" sParam="" /> <!-- Apply the 5th STYLE --> <Action type="0" message="2306" wParam="0" lParam="0" sParam="" /> <!-- Hit the RIGHT key --> <Action type="0" message="2454" wParam="0" lParam="0" sParam="" /> <!-- Select to START of line --> <Action type="0" message="2180" wParam="0" lParam="0" sParam="" /> <!-- Hit on the DELETE key --> </Macro>-
Then save the modifications (
Ctrl + S) and closeshortcuts.xml(Ctrl + W) -
Now, stop and re-start N++
-
Open any file containing some
ADNsequences -
If some
ADNsequences contained lower-cased letters :-
Open the Find dialog, with
Search > Find...(Ctrl + F) -
Untick, if necessary, the
Match caseoption -
Hit the
ESCkey to close the Find dialog
-
-
Execute this macro, running the command
Macro > ADN Test, which should be visible, along with your present macros, if any ( You may assign a shortcut, as well )
Et voilà ;-))
Best Regards,
guy038
-
-
@guy038 always tries out my proposed solutions to problems, so this time I tried @guy038 's solution and it works nicely. First I obtained some data to try it on from here. Then, executing the macro on the data resulted in this nicely visual view:
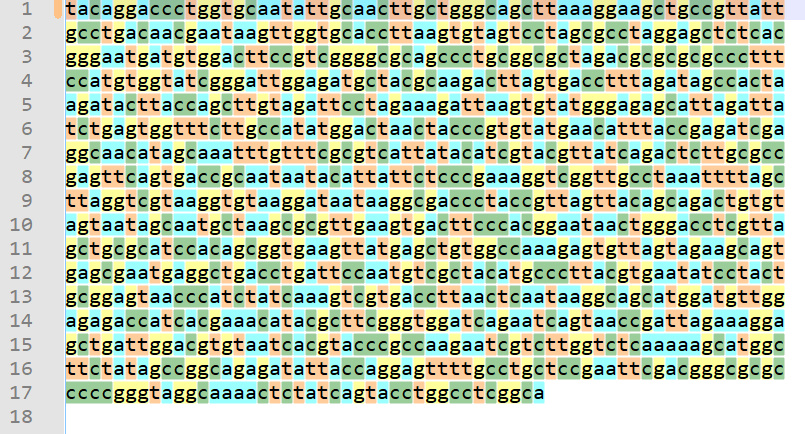
Perhaps the only note to make is that, upon any future change to
shortcuts.xmlthat is made automatically by Notepad++, will cause @guy038 's nice comments on each line of the macro to be removed! :-( -
To give @Ekopalypse fair time as well, I decided to try the lexing script.
Here’s what I did:
-
Create a barebones UDL called
ADNTest. I changed/added nothing, just saved an “empty” definition. -
Get the latest version I could ascertain of the EnhanceUDLLexer.py file. [Sadly, the thread linked to (above by Peter) for this starts out with
I opened this thread just to inform you that I have reworked the script a little bit. Such an opening is begging for a link to an original discussion thread!] -
Modify that file as follows:
-
lexer_name = 'ADNTest' -
Comment out existing
regexesvariable settings (from the Examples section) and add in the following ones (using some “random” color numbers):
regexes[(0, (252, 173, 67))] = (r'a', 0) regexes[(1, (152, 73, 167))] = (r'g', 0) regexes[(2, (52, 37, 176))] = (r'c', 0) regexes[(3, (25, 17, 77))] = (r't', 0)-
Run the script.
-
Generate some random DNA data from the link provided earlier by me into a new Notepad++ tab.
-
Change the Language of the tab to be my
ADNTestUDL.
It worked nicely, giving me this view of the data:

Overall, perhaps I give the nod to @guy038’s solution because it is a lot simpler for someone to use that isn’t already using Pythonscript.
-
-
-
@Alan-Kilborn said in UDL syntax highlighting: non-delimited keywords (for nucleotide sequences):
@guy038 is dyslexic – no
Actually, @guy038 is French. “DNA”:“Deoxyribonucleic acid”::“ADN”:“acide désoxyribonucléique”, per a quick google search.
-
@Alan-Kilborn said in UDL syntax highlighting: non-delimited keywords (for nucleotide sequences):
regexes[(0, (252, 173, 67))] = (r’a’, 0)
regexes[(1, (152, 73, 167))] = (r’g’, 0)
regexes[(2, (52, 37, 176))] = (r’c’, 0)
regexes[(3, (25, 17, 77))] = (r’t’, 0)I finally gave in and actually tried using the script. It does work fairly easily, like you showed.
I confirmed with the following data that it is case sensitive.
gattaca acctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa GATACCGCTGGACCTAAAAGCGAGGGGACTTCTGGGCGGCGTATCCAA GATACCGCTGGACCTAAAAGCGAGGGGACTTCTGGGCGGCGTATCCAA GATACCGCTGGACCTAAAAGCGAGGGGACTTCTGGGCGGCGTATCCAA GATACCGCTGGACCTAAAAGCGAGGGGACTTCTGGGCGGCGTATCCAA GATACCGCTGGACCTAAAAGCGAGGGGACTTCTGGGCGGCGTATCCAA GATACCGCTGGACCTAAAAGCGAGGGGACTTCTGGGCGGCGTATCCAA GATACCGCTGGACCTAAAAGCGAGGGGACTTCTGGGCGGCGTATCCAA gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggacctaaaagcgaggggacttctgggcggcgtatccaa gataccgctggThus,
regexes[(0, (252, 173, 67))] = (r'[Aa]', 0) regexes[(1, (152, 73, 167))] = (r'[Gg]', 0) regexes[(2, (52, 37, 176))] = (r'[Cc]', 0) regexes[(3, (25, 17, 77))] = (r'[Tt]', 0)will use Alan’s same colours, but allow lower case or upper case.
To make it automatic, I added the following to the
startup.py(by Plugins > Python Script > Scripts, thenCtrl+Clickonstartup)# to automatically import eko's EnhanceUDLLexer: d = notepad.getPluginConfigDir() + '\\PythonScript\\scripts\\' sys.path.append(d) import EnhanceUDLLexer EnhanceUDLLexer.EnhanceUDLLexer().main()When you exit and reload Notepad++, DNA files should now automatically be colored.
However, as Alan said, @guy038’s macro is probably easier to use for someone who doesn’t already use PythonScript (or for anytime that Notepad++ updates break PythonScript notifications/callbacks).
-
Hello, @n-iels, @alan-kilborn, @peterjones and All,
Thanks, Alan, for your kind words ! Yes, Peter is just right about it :
ADN( “Acide désoxyribonucléique” ) is simply the French initialism forDNA( “Deoxyribonucleic acid” ). So, I’m not thinking, yet, as native English-American people ? My bad ;-))By the way, do you know the difference between an acronym and an initialism ?
Acronym Signification Syntactic details Pronunciation ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ GIF Graphics Interchange Format Pure acronym, formed from initial letters As a SINGLE word Radar Radio Detection And Ranging Pure acronym, formed from both initial and non-initial letters As a SINGLE word CD-ROM Compact Disc Read-Only Memory Both, acronym and initialism, formed from initial letters _C_ _D_ _Rom_ DNA DeoxyriboNucleic Acid Pure initialism, formed from both initial and non-initial letters _D_ _N_ _A_ HTML HyperText Markup Language Pure initialism, formed from both initial and non-initial letters _H_ _T_ _M_ _L_ VHF Very High Frequency” Pure initialism, formed from initial letters _V_ _H_ _F_ From : https://en.wikipedia.org/wiki/Acronym#Nomenclature and : https://en.wikipedia.org/wiki/Acronym#Comparing_a_few_examples_of_each_type
Now, regarding the macro, I simply used the default N++ colors of the Mark styles. I just avoided the
4thstyle, which seems too dark purple, to my mind ! Of course, all these style colors can be changed inSettings > Style Configurator... > Global styles > Mark Style #1 to #5;-))Best Regards,
guy038
-
@guy038 said in UDL syntax highlighting: non-delimited keywords (for nucleotide sequences):
do you know the difference between an acronym and an initialism
I don’t. I still don’t. And I don’t know why it is important enough for their to be a distinction. :-)
-
Hi, @alan-kilborn and All,
When I was elaborating my answer to you, about the
"ADN -DNA"difference, I thought , of course, about the French name acronyme to describe these things, with the intention of choosing the correct English word, afterwards, which I would had inserted in my post.So, after a search on wikipedia.fr I was surprised to learn that different kinks of acronyms exist. Particularly, the two main families :
- True acronyms, ( French word : acronymes ), which are pronounced as a single word
- Pseudo acronyms,called initialisms ( French word : sigles ), which are pronounced as individual letters
For instance, in the Wikipedia article, it it said :
… some dictionaries and usage commentators define acronym to mean an abbreviation that is pronounced as a word, in contrast to an initialism (or alphabetism)—an abbreviation formed from a string of initials (and possibly pronounced as individual letters)…
So, it’s out of curiosity that I noticed these differences. Just consider that @guy038’s mind was rather creating a useless story ;-)) And, of course, absolutely no relation at all with Notepad++ and @n-iels problem ;-))
Last remark :
I have often noticed that people who know several languages are more respectful of their own language ! For example, I don’t like the “Frenglish” terms. In my opinion, it’s just an honor to speak and write well in each language ! Of course, I do understand that no language is really fixed and everyone evolves on their own ;-))
Best Regards
guy038
-
Thanks, you are amazing… I have now tried both solutions:
The Python script by @Ekopalypse works nicely, but has the following problems:
- It slows Notepad++ down dramatically, to the point that working with a medium-size sequence file (500 k chars, mostly dashes) becomes impossible.
- The script does not allow you to style the background. But background styling is what we usually want for this purpose.
- Also, I noticed, that you cannot invoke the script twice (e.g. after changing colors).
BTW, the standard colors that I prefer would be:
regexes[(0, (224, 0, 0))] = (r'[tTuU]', 0) # red regexes[(1, ( 2, 140, 0))] = (r'[aA]' , 0) # green regexes[(2, ( 0, 0, 230))] = (r'[cC]' , 0) # blue # regexes[(3, ( 0, 0, 0))] = (r'[gG]' , 0) # black (default)As for the macro by @guy038:
- It is much faster and actually makes editing the sequence file from above possible.
- But on one of my computers, the A’s are not styled (I still have the standard colors set in the style configurator).
- Understanding and editing the macro code is almost impossible and I could not find a useful macro documentation that is online (most links point to the now defunct npp wiki). It would be nice to have the same algorithm in Python Script…
- The macro is not dynamic, i.e. after editing your sequences, you have to manually re-invoke that macro. Until then, your new sequence is styled wrong or not at all.
Both solutions allow me to finish my current project (thanks again!). But the preferable solution for non-hackers would be to make this a standard language (“Nucleotides”) after enhancing the UDL highlighter as discussed on the bug tracker: https://github.com/notepad-plus-plus/notepad-plus-plus/issues/7622.
-
Thank you very much for the testing and feedback of the test results.
I’m afraid your use case reaches the limits of the script.
Since the script only colors the current visual area, you seem to have very long lines with many hits, which leads to delays.
The script is currently using, as you mentioned earlier, the
textforeground indicator, this could be changed to get the same format as Guy’s solution.
A restart of the script can be accomplished by deleting the class EnhanceUDLLexer via the PythonScript Console,
withdel EnhanceUDLLexerand another start.Can you tell me how much data you currently have, i.e. how long individual lines are in the visual area?
I’d like to see if it’s possible to optimize the script, but I’m afraid, as I’ve said, this probably won’t do much good.But I understand, that a native support would be the preferred solution.
-
@N-iels said :
It slows Notepad++ down dramatically, to the point that working with a medium-size sequence file (500 k chars, mostly dashes) becomes impossible.
@Ekopalypse said :
Since the script only colors the current visual area, you seem to have very long lines with many hits, which leads to delays.
@Ekopalypse Deja vu? :-)
-
@N-iels said in UDL syntax highlighting: non-delimited keywords (for nucleotide sequences):
I could not find a useful macro documentation that is online
There is an overview of recording macros in the official documentation at https://npp-user-manual.org/docs/macros/, and a more-detailed description of the
<Macros>section ofshortcuts.xmlat https://npp-user-manual.org/docs/config-files/#macros, with links to the list of Notepad++ and Scintilla messages that the<Action>tags can refer to. -
hehe :-) I guess :-D
Sometimes rectangles might be useful :-)) (@all sorry, that’s an insider) -
@N-iels said :
Understanding and editing the macro code is almost impossible
TBH, most people never attempt to edit it, except maybe to slightly tweak it, or to do something that is not normally possible. What people do is to record their actions with the macro recorder. @guy038 probably did this when deriving his solution, followed possibly by some hand-tweaking.
It would be nice to have the same algorithm in Python Script
I think the macro solution is a good one. While it would be possible to duplicate the functionality with Pythonscript, I don’t know that it is worth the effort, except maybe for instructional (how would one do it) purposes…hmmm…
-
@Alan-Kilborn said in UDL syntax highlighting: non-delimited keywords (for nucleotide sequences):
except maybe for instructional (how would one do it) purposes
Beat you to it. :-)
# encoding=utf-8 """in response to https://notepad-plus-plus.org/community/topic/18512/ Convert this MACRO into PythonScript <Macro name="ADN Test" Ctrl="no" Alt="no" Shift="no" Key="0"> <Action type="2" message="0" wParam="43032" lParam="0" sParam="" /> <!-- DELETE ALL styles --> <!-- NPPM_ --> <Action type="0" message="2453" wParam="0" lParam="0" sParam="" /> <!-- Go to START of line --> <!-- SCI_ NoString --> <Action type="1" message="2170" wParam="0" lParam="0" sParam="A" /> <!-- Write the letter A --> <!-- SCI_ YesString --> <Action type="1" message="2170" wParam="0" lParam="0" sParam="T" /> <!-- Write the letter T --> <!-- SCI_ YesString --> <Action type="1" message="2170" wParam="0" lParam="0" sParam="G" /> <!-- Write the letter G --> <!-- SCI_ YesString --> <Action type="1" message="2170" wParam="0" lParam="0" sParam="C" /> <!-- Write the letter C --> <!-- SCI_ YesString --> <Action type="0" message="2453" wParam="0" lParam="0" sParam="" /> <!-- Go to START of line --> <!-- SCI_ NoString --> <Action type="0" message="2307" wParam="0" lParam="0" sParam="" /> <!-- Select the NEXT char --> <!-- SCI_ NoString --> <Action type="2" message="0" wParam="43022" lParam="0" sParam="" /> <!-- Apply the 1st STYLE --> <!-- NPPM_ --> <Action type="0" message="2306" wParam="0" lParam="0" sParam="" /> <!-- Hit the RIGHT key --> <!-- SCI_ NoString --> <Action type="0" message="2307" wParam="0" lParam="0" sParam="" /> <!-- Select the NEXT char --> <!-- SCI_ NoString --> <Action type="2" message="0" wParam="43024" lParam="0" sParam="" /> <!-- Apply the 2nd STYLE --> <!-- NPPM_ --> <Action type="0" message="2306" wParam="0" lParam="0" sParam="" /> <!-- Hit the RIGHT key --> <!-- SCI_ NoString --> <Action type="0" message="2307" wParam="0" lParam="0" sParam="" /> <!-- Select the NEXT char --> <!-- SCI_ NoString --> <Action type="2" message="0" wParam="43026" lParam="0" sParam="" /> <!-- Apply the 3rd STYLE --> <!-- NPPM_ --> <Action type="0" message="2306" wParam="0" lParam="0" sParam="" /> <!-- Hit the RIGHT key --> <!-- SCI_ NoString --> <Action type="0" message="2307" wParam="0" lParam="0" sParam="" /> <!-- Select the NEXT char --> <!-- SCI_ NoString --> <Action type="2" message="0" wParam="43030" lParam="0" sParam="" /> <!-- Apply the 5th STYLE --> <!-- NPPM_ --> <Action type="0" message="2306" wParam="0" lParam="0" sParam="" /> <!-- Hit the RIGHT key --> <!-- SCI_ NoString --> <Action type="0" message="2454" wParam="0" lParam="0" sParam="" /> <!-- Select to START of line --> <!-- SCI_ NoString --> <Action type="0" message="2180" wParam="0" lParam="0" sParam="" /> <!-- Hit on the DELETE key --> <!-- SCI_ NoString --> </Macro> https://www.bioinformatics.org/sms2/random_dna.html catctaaagggattagttcctgccctcatattcactatccgacccctttaactgtgatgt cctcgctttttctcgtgagagctgtgaatctttgtgccgtttccaacaaggcctggagcc ttttcaatgcttgagggtttcaccgcgggtctaacggatgctaagaaaggggtgcggagg aagggtctttatgctggccgtcggcggttgagagctctgacctataccatggatcccgcg agcgcggttacgggcaataagggcctcactatgcctcgaacacattgtggacaaagtgta gtcgaacccacacacgcgcgagactttagggtgtcgaacagtaccatctaattgatggga agaaatggtttcgtaccacccccgtcgctcagcttagacgggccagagaggggatgggtg gtcagtggcgtcggttggtgaccgtagaattcgttacagagcgatgttgtatagcttttt agacgtaggctagcgttttaacttctacaactccagtgattgggttgatggtctgtttgc ttaccagtcaggtcagctcccgctcatggttctctcgcaaattacttggtcacaccgtga aagctccacgcaaactaatagtgggattctacactaaagggcgtcactatcacttcttat acattatagacgtaactacagtagacatactcgcaagcccgctaacgggagcacagatgt tgagggtatcagcttctgcgactcgggctggatccgatatttttatgcaatgcatctgag actggcctccctgctacctctacggaagctggtacgaagcgcgctgccttcgactgaaac ttgcatgcataagttaatgtagtgcagcgcaggtcagccaacataagtagtgagcccagc cgctggcaggacagttgtcgcggtaaatcacacgtgtggtgaccatctccccatttacag gtgttagaaaagcaacttcgtattaatccattaatctgag """ notepad.menuCommand(43032) # <!-- DELETE ALL styles --> IDM_SEARCH_CLEARALLMARKS = Search > Unmark All > Clear All Styles editor.vCHomeWrap() # <!-- Go to START of line --> editor.replaceSel("A") # <!-- Write the letter A --> editor.replaceSel("T") # <!-- Write the letter T --> editor.replaceSel("G") # <!-- Write the letter G --> editor.replaceSel("C") # <!-- Write the letter C --> editor.vCHomeWrap() # <!-- Go to START of line --> editor.charRightExtend() # <!-- Select the NEXT char --> notepad.menuCommand(43022) # <!-- Apply the 1st STYLE --> Search > Mark All > Using 1st Style editor.charRight() # <!-- Hit the RIGHT key --> editor.charRightExtend() # <!-- Select the NEXT char --> notepad.menuCommand(43024) # <!-- Apply the 2nd STYLE --> Search > Mark All > Using 2nd Style editor.charRight() # <!-- Hit the RIGHT key --> editor.charRightExtend() # <!-- Select the NEXT char --> notepad.menuCommand(43026) # <!-- Apply the 3rd STYLE --> Search > Mark All > Using 3rd Style editor.charRight() # <!-- Hit the RIGHT key --> editor.charRightExtend() # <!-- Select the NEXT char --> notepad.menuCommand(43030) # <!-- Apply the 5th STYLE --> Search > Mark All > Using 5th Style editor.charRight() # <!-- Hit the RIGHT key --> editor.vCHomeWrapExtend() # <!-- Select to START of line --> editor.clear() # <!-- Hit on the DELETE key --> # use notepad.menuCommand(43032) or Search > Unmark All > Clear All Styles to clear the styles when done